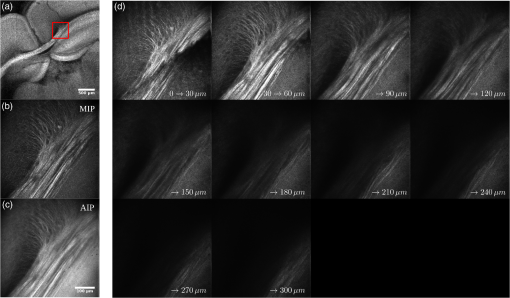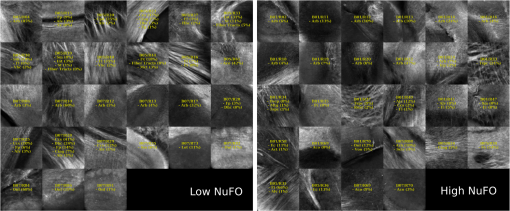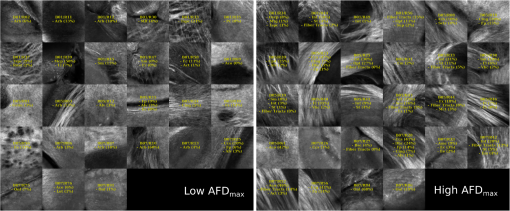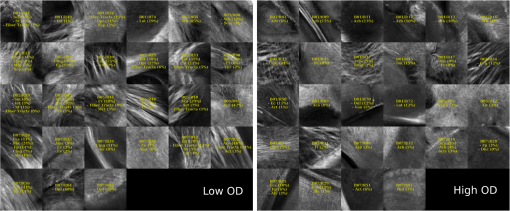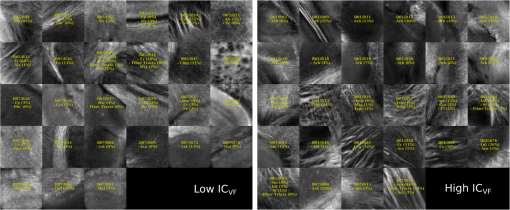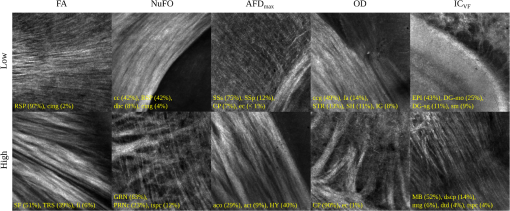
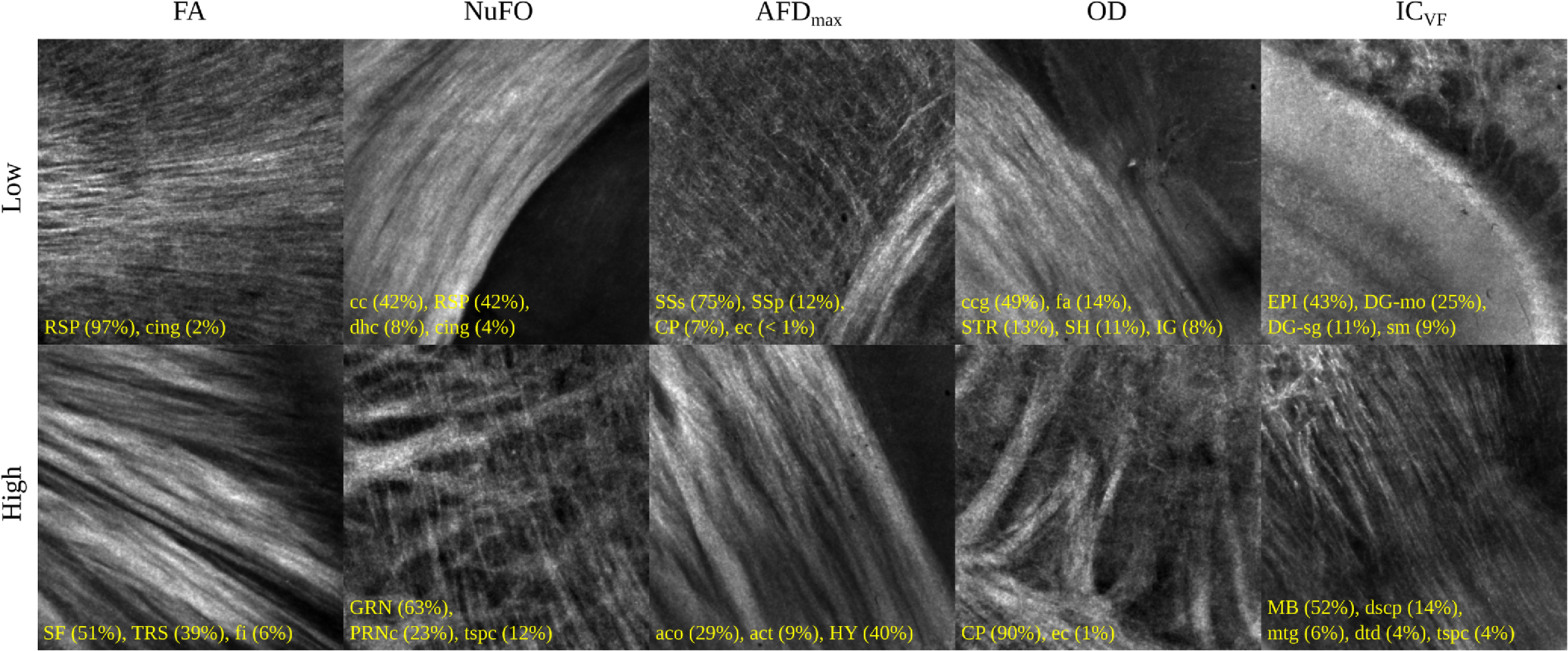
Examples of 40× ROIs associated with low- and high-dMRI measure values. Each image spans an FOV of 0.5×0.5 mm2 and is an AIP over 250 μm. The yellow annotations indicate the brain structures and their volume fraction within the ROIs. The acronyms follow the Allen mouse brain convention. RSP, retrosplenial area; cing, cingulum bundle; SF, septofimbrial nucleus; TRS, triangular nucleus of septum; fi, fimbria; cc, corpus callosum; dhc, dorsal hippocampal commissure; GRN, gigantocellular reticular nucleus; PRNc, pontine reticular nucleus caudal part; tspc, crossed tectospinal pathway; SSs, supplemental somatosensory area; SSp, primary somatosensory area; CP, caudoputamen; ec, external capsule; aco, anterior commissure olfactory limb; act, anterior commissure temporal limb; HY hypothalamus; ccg, genu of the corpus callosum; fa, corpus callosum anterior forceps; STR: striatum; SH, septohippocampal nucleus; IG, induseum griseum; EPI, epithalamus; DG-mo, dentate gyrus-molecular layer; DB-sg, dentate gyrus-granule cell layer; sm stria medullaris; MB, midbrain; dscp, superior cerebellar peduncle decussation; mtg, mammilotegmental tract; dtd, doral tegmental decussation; and tspc, crossed tectospinal pathway.
|